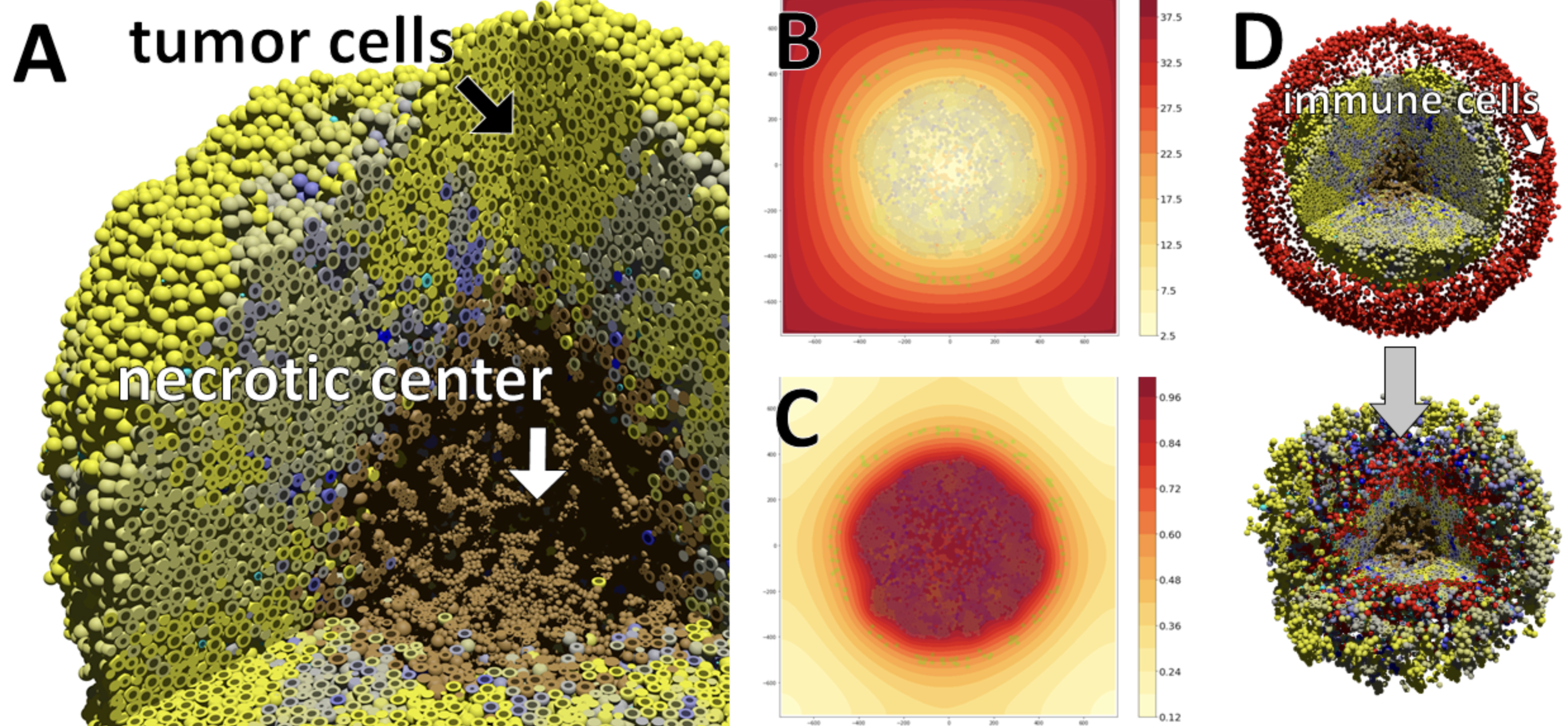
Project Abstract:
In this paper, we utilize OpenACC, a directive-based programming model to accelerate the diffusion portion PhysiCell, a cross-platform agent-based biosimulatione work that has been adopted in cancer, infectious diseases and other complex biological problems. Using NVIDIA HPC SDK OpenACC 21.3, we demonstrate an almost 40x speedup using managed memory on the state-of-the-art NVIDIA Ampere 100 (A100) GPU compared to a serial AMD EPYC core 7742 for a 360 simulated minutes input dataset. We also demonstrate 9x speedup on the 64 core AMD EPYC 7742 multicore platform using NVIDIA HPC SDK OpenACC 21.3. By using OpenACC for both the CPUs and the GPUs, we maintain a single source code base, thus creating a portable yet performant solution.